 |
|
 |
|
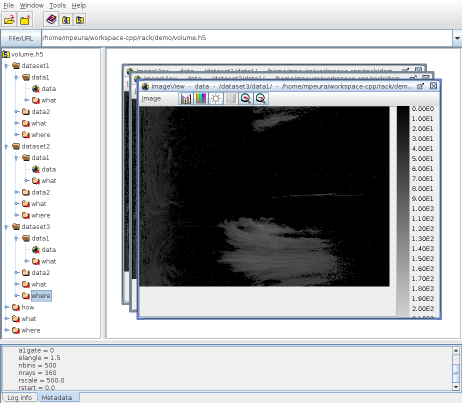
Rack supports reading sweep and volume files in HDF5 format using OPERA information model (ODIM). In addition, supports reading and writing data as images and text files.
Input files are given as plain arguments or alternatively with explicit command --inputFile , abbreviated -i. If several files are given, they will be internally combined in the internal HDF5 structure, adding datasets incrementally.
Outputs are generated using --outputFile , abbreviated -o. Hence, combining sweep files to a volume is obtained simply with
In combining sweep data to volumes, Rack creates and updates one /dataset<i> group for each elevation angle . Further, Rack creates and updates one /data<i> group for each quantity ; new input overwrites existing data of the same quantity.
If quality information – stored under some /quality[i] group and marked with what:quantity=QIND – is read, the overall quality indices are updated automatically. See Reading and combining quality data below for further details.
Using --outputFile , the structure of the current data can be written as plain text in a file (*.txt) or to standard input (-):
File volume.txt will consist of lines of ODIM entries as follows:
Note that strings are presented in double quotes and arrays as comma-separated values in brackets.
Based on metadata, it is often handy to automatically compose filenames. This is possible using --expandVariables as follows.
For using format templates and hadling multiple files on a single command line, see Formatting metadata output using templates.
Volume can be also created using a text file like the one created above, that is, with the desired structure and data types.
One can modify the metadata directly from the command line by means of --setODIM <path>:<attribute>=<value> command.
The command has a special shorthand --/<path>:<attribute>=<value> . For example:
This feature can be used for completing incomplete ODIM metadata or adding arbitrary metadata. The actual radar data can be read or saved as image files as explained below.
In addition to HDF5 format, Rack support three image formats:
When writing data with --outputFile (-o) command, the applied image format is determined from the filename extension. By default, the first dataset encountered in the internal data structure is selected, hence typically /data1/dataset1/data . The source can be changed with --select command.

-o .Metadata (ODIM variables) are written in the comment lines (PGM and PNM). The syntax follows that of a text dump produced with --outputFile as explained above with volume.txt , but only the last path component (what , where, how) is included. For example, a resulting header of a PGM file - here pruned for illustration - looks like:
The comments can be overriden with --format command. This experimental feature covers currently:
'{' , it is read in as JSON , and (first-level) key-value pairs are stored as GDAL attributes.In addition, for GeoTIFF files, attributes set with --/how:GDAL:<key>=
will be also added as GDAL variables.
Rack can also save all the data sets in separate files with a single command, producing sweep000.png , sweep001.png and so on. This is achieved with --outputRawImages , abbreviated -O . The command stores the image data directly, without rescaling pixel values.
Reading image files creates and updates internal HDF5 structure, adding grid data:
/data<i> or /quality<i> group containing an empty /data (ie. uninitialized image), or if not found:/data<i>/data of the existing /dataset<j> with the highest index j Metadata can be set
what:quantity="DBZH" ) --/dataset1/data2/what:quantity=DBZH (see changing)Metadata can be set before or after image inputs. Example:
Some important ODIM attributes can be added automatically with --completeODIM command, which sets nbins , nrays , xsize , and ysize equal to data dimensions, if already loaded as image.
Rack produces GeoTIFF images under the following limitations:
scale and offset ).gdalinfo to check outputepsg:3844, epsg:3035, and epsg:3995) raise errors – see Cartesian conversions and composites for details.display ) produce rendering errors of 16-bit images having width not multiple of tile width
Experimental. The current data structure can be written to a HTML file which displays the data in a clickable tree. The data arrays are stored in PNG files in sub directory named with the basename of the output file. The structure of the directory repeats the hierarchy of the original HDF5-ODIM data.
Examples:
Example:
The resulting file consists of lines containing values and counts
Example:
The contents of hierarchical radar data can be displayed in a simple tree format using outputTree command or by changing file extension to .tre when using the general command -o / --outputFile .
Rack can be also used to output the tree-like hierarchy of radar data in dot format (http://www.graphviz.org/documentation/). From that format, one may use Graphviz dot program to produce tree graphs in various image formats link png , pdf and svg. For example:
The desired path can be selected with --select path=<group>[/<group>] or simply --select <group>[/<group>] because path is the first argument key.
The most relevant groups are dataset: , and data: , and quality: . The colon : is compulsory for separating data<index> groups from lower level data group in ODIM. In the current version, groups what , what and how are handled somewhat automagically. (See Selecting data .)

dot file output.In an operative environment, two parallel processes may perform quality control. Rack intelligently combines the resulting files using the following logic:
quantity=QIND ) is updated, based on either target class field (quantity=CLASS) or (quantity=<class-name> ):quantity=CLASS) is provided, QIND and CLASS will be updated directly from those.CLASS is not provided, QIND will be updated from the specific class probability (quantity=<class-name> ).QIND (typically /quality1 ), how:task_args will be updated by the names of the added classes.In the following example, the quantities (detection classes) applied are those produced by Rack but could be thought as if they were produced by some other software, with same or similar names.
Consider two quality control processes, the first producing detection of EMITTER and JAMMING , stored in file volume-det1.h5 , containing following data (among others):
Assume the other process detecting SHIP and SPECKLE stored in volume-det2.h5 , respectively:
These files can be combined simply with
The resulting file contains structure as follows:
Just like after running several detection processes, after reading files containing detection fields, global ie. elevation-specific quality data (QIND and CLASS ) are not implictly combined to local ie. quantity-specific quality data. As explained in detection , this combination takes place automatically if any removal command or --c –aQualityCombiner command is issued.
 1.9.8
1.9.8